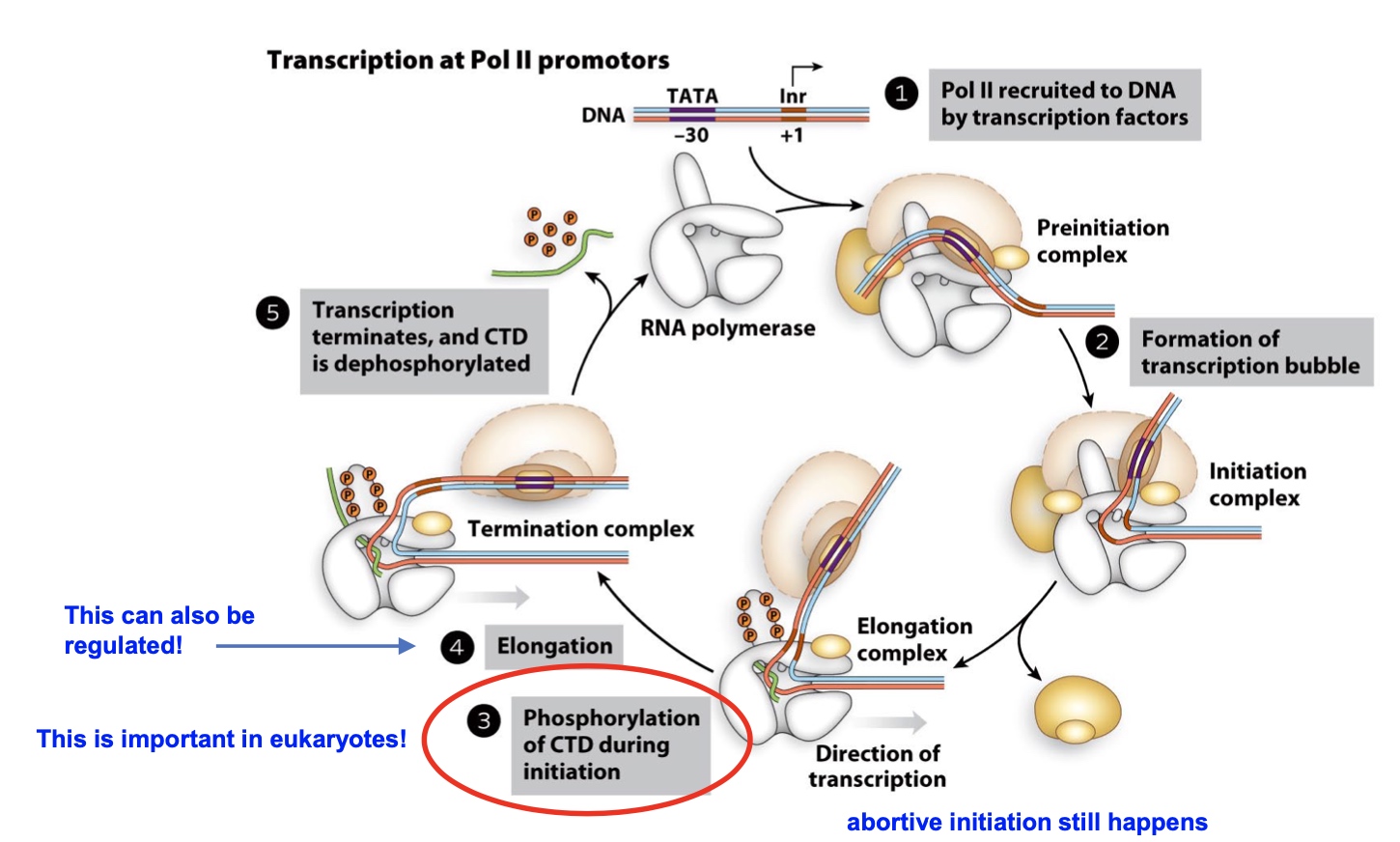
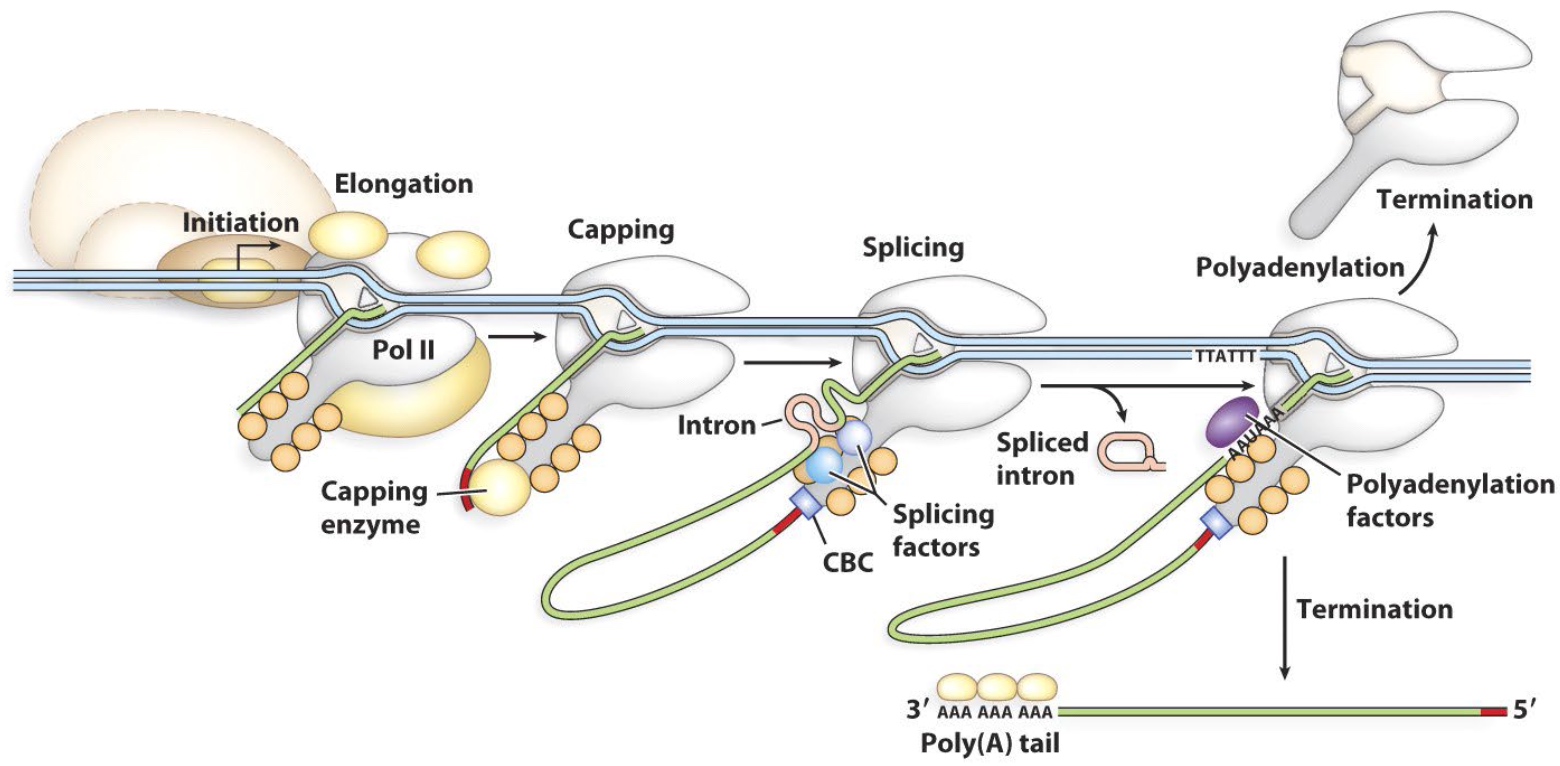
Overview of eukaryotic Pol II transcription cycle
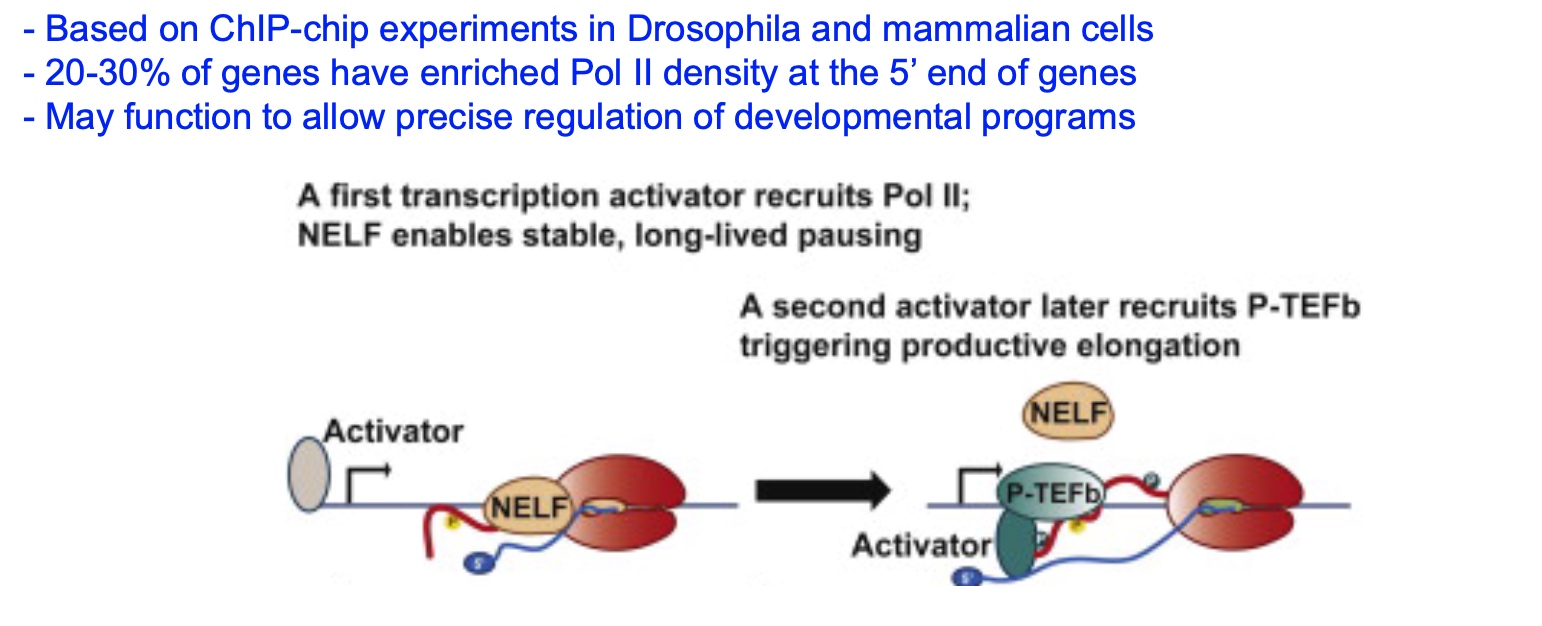
Eukatyotic "Promoter Proxial Pausing"
- NELF – negative elongation factor Binding to RNA pol II causes it to stably pause 30-50nt downstream of txn. start site
- P-TEFb – positive transcription elongation factor Signaling pathways activate it. Transcription elongation resumes
Two Models for eukaryotic transcription termination
- Allosteric – RNA structure causes termination (like rho-independent)
- Torpedo – RNA pol is forced off the template by Xrn2 exonuclease (like rho-dependent)
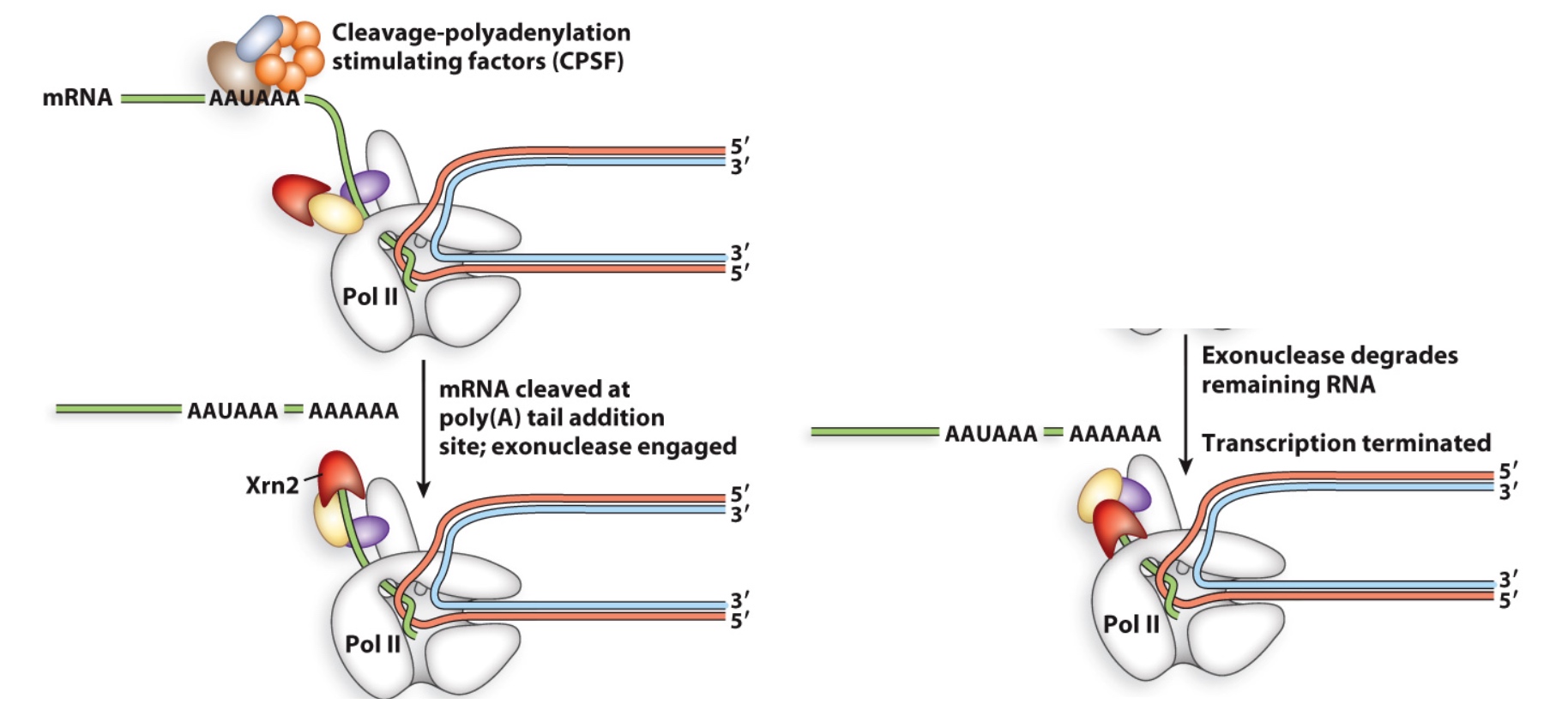
CPSF(Cleavage-polyadenylation stimulating factors) will bind to "AAUAAA", then it will cut the polyA tail addition site.
Questions
For these eukaryotic transcription steps, state which of the two steps happens FIRST.
- A. binding of TFIID to the promoter / binding of RNA pol to the promoter
- B. binding of NELF to RNA pol II / promoter clearance
- C. dissociation of RNA pol II from the DNA template / cleavage of the mRNA by an endonuclease
A: Initiation
B: Regulation
C: Termination
Several processes can synergize to regulate transcriptional activation
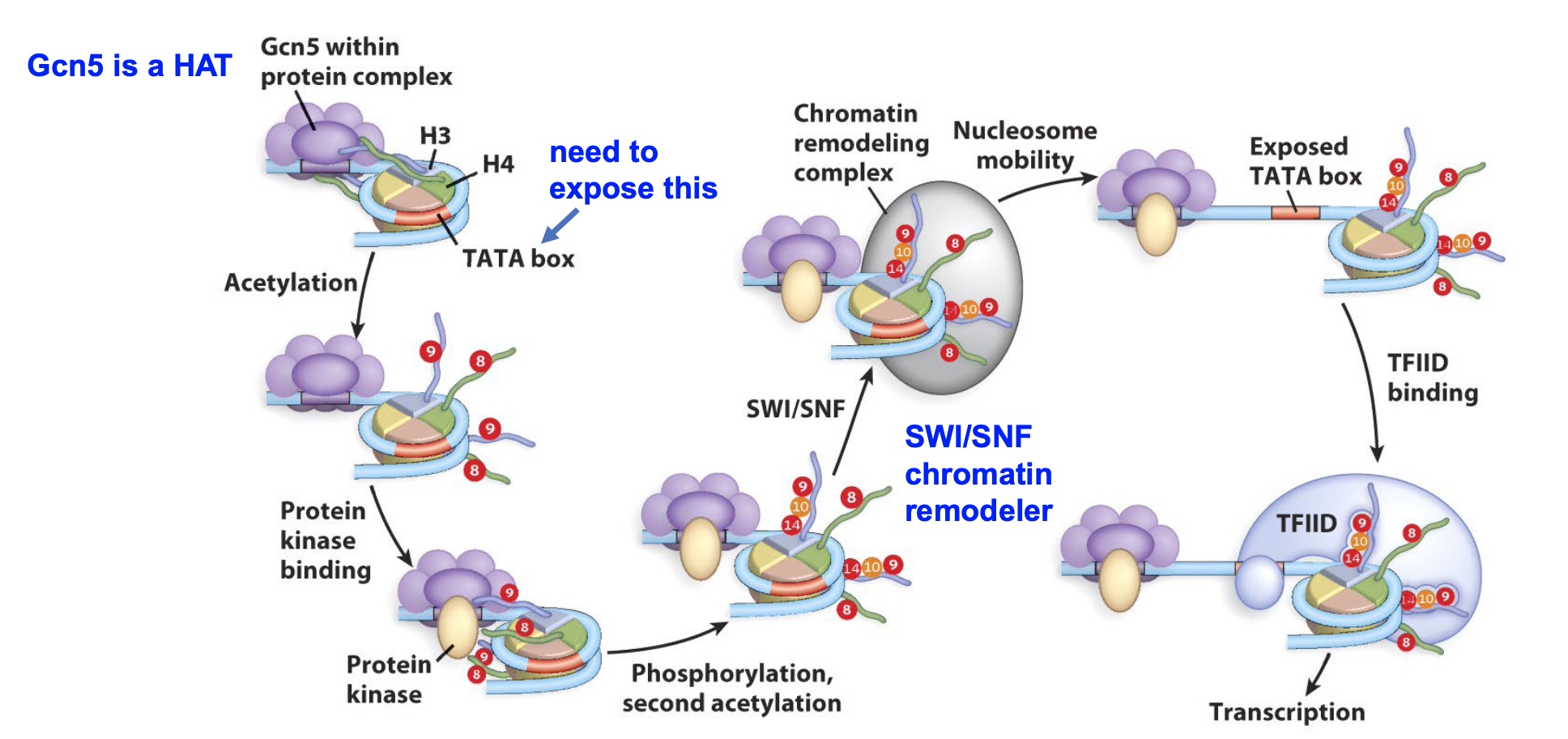
Once the TATA bos is exposed, TFIID come to bind the promotor and then recruit RNA Pol to come and start transcription.
DNA methylation also represses gene expression
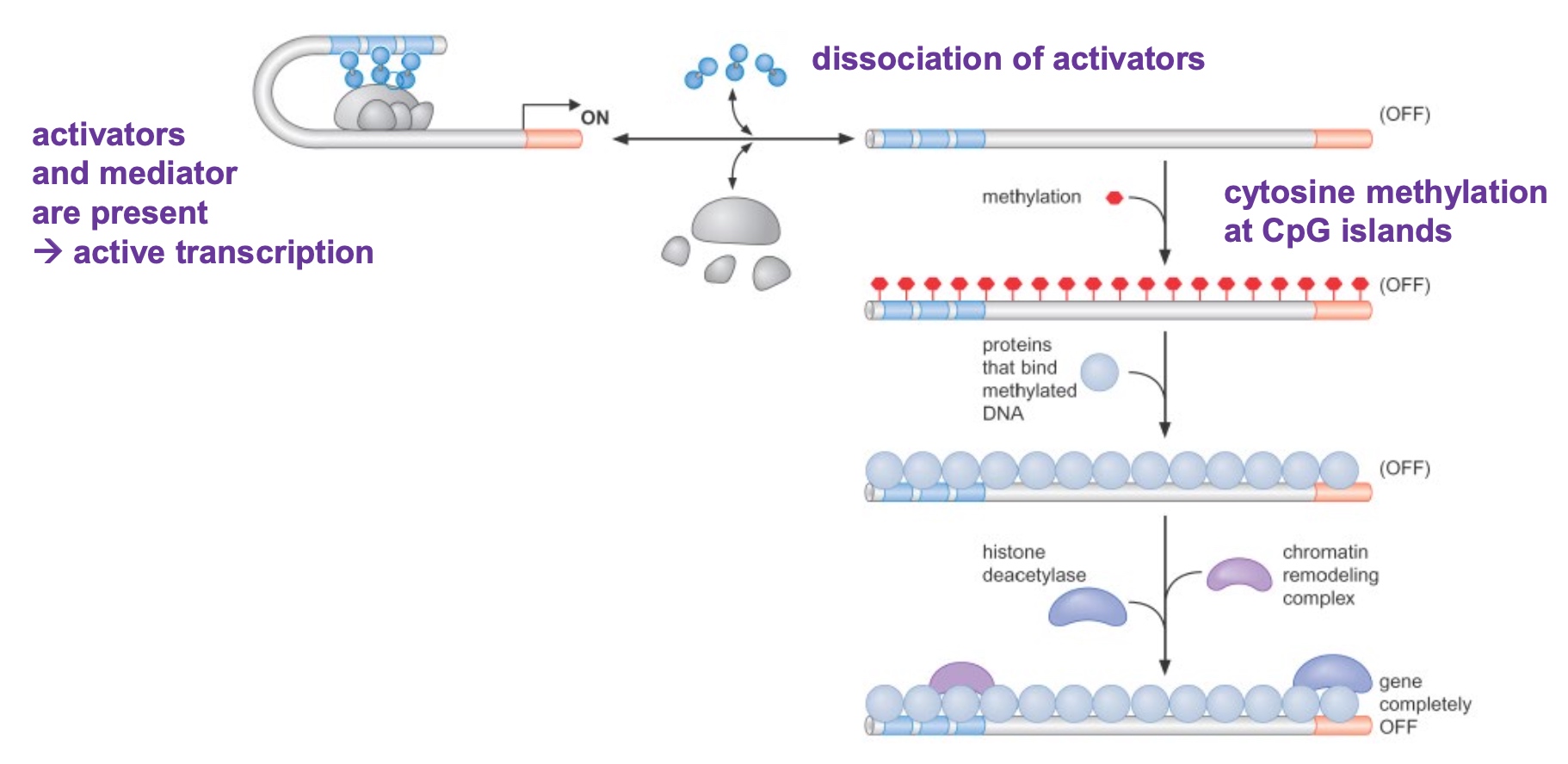
Cytosine methylation usually repress the gene repression.
Poll Everywhere Question
The p53 gene is a tumor suppressor gene. Assuming that the p53 gene is normal (not mutated), which of the following would you expect to be true about the promoter of the p53 gene in cancer cells?
- A. DNA is methylated
- B. DNA is unmethylated
- C. histones are acetylated
- D. histones are deacetylated
- E. nucleosome positioned at the TATA box
- F. nucleosomes absent from the TATA box
Methylated??
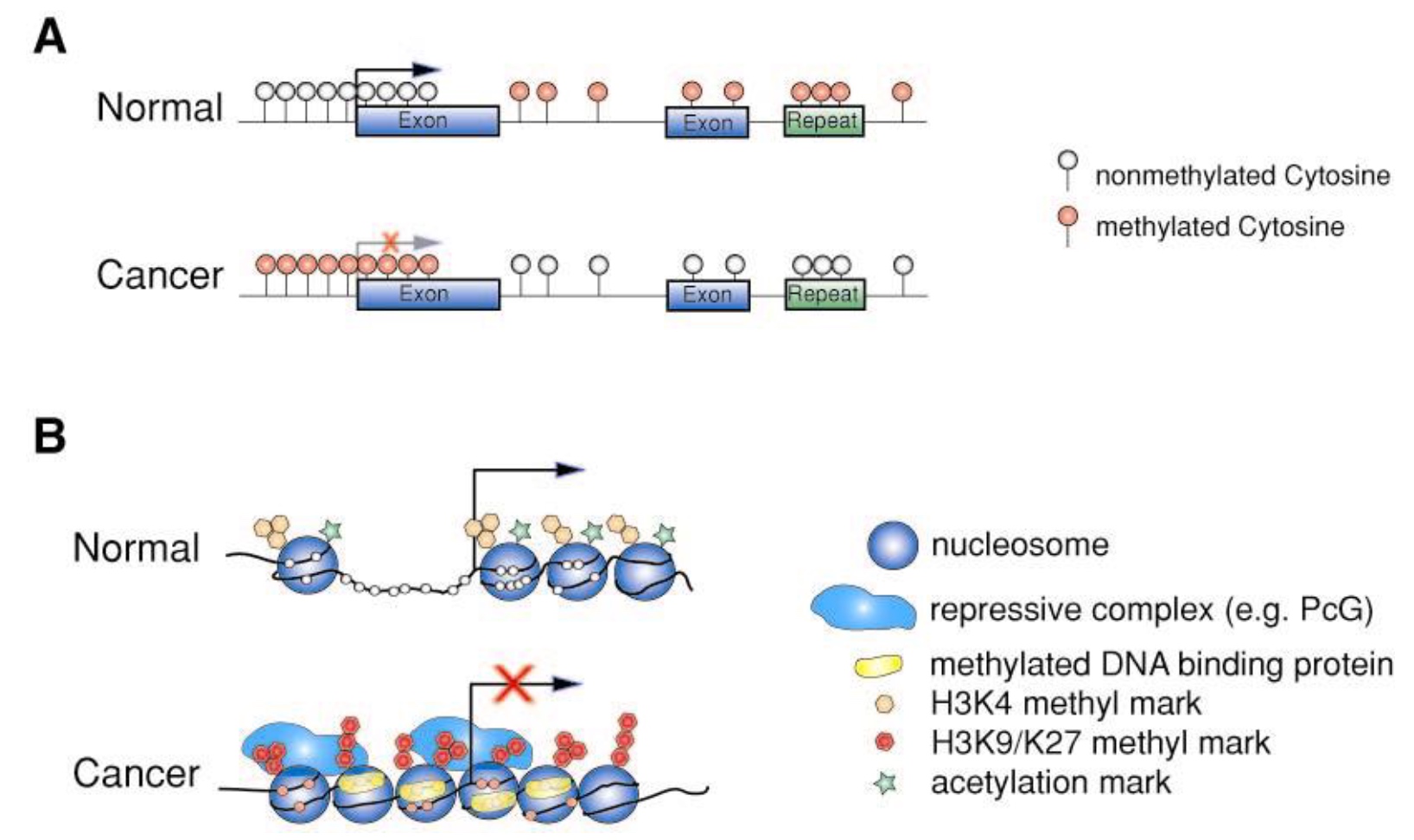
The chromatin context at promoters of many genes is different in normal vs. cancer cells
Transcription nomenclature - review

- Will the promoter be included in the pre-mRNA?
- No.
- What parts will be included in the processed mRNA?
- Cap, Exon1, Exon2 and PolyA tail. mRNA will do capping, splicing and polyA tailing.
- Where do you find the 5’ and 3’ untranslated regions (UTRs)?
RNA processing in eukaryotes: RNA pol CTD coordinates transcription and pre-mRNA processing
TTATTT -- AAUAAA
Time course of an in vitro splicing reaction

Why intron and intron-exon2 move slower than pre? Because there are circle on them.

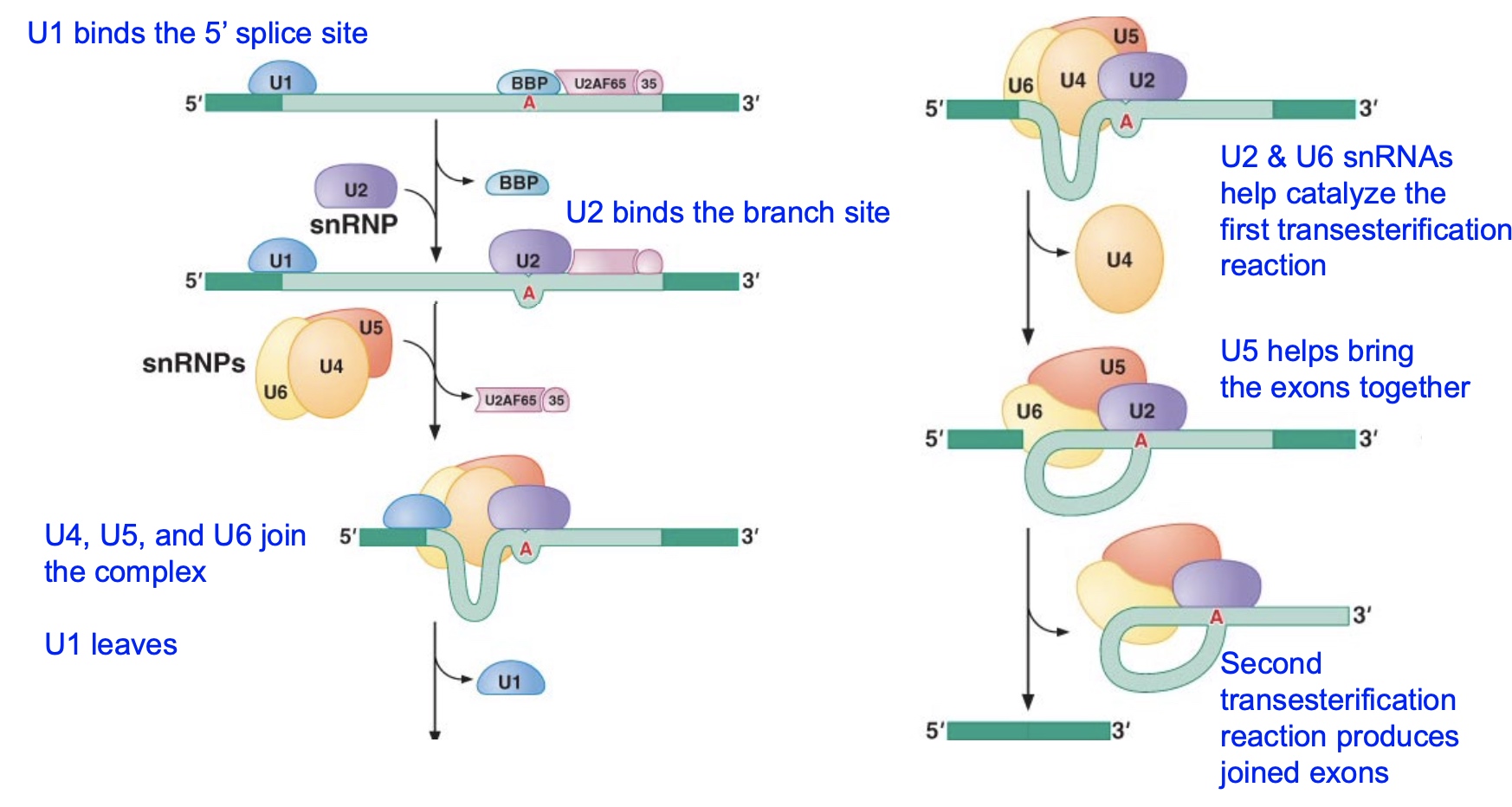
Overview of the spliceosome-mediated splicing reaction
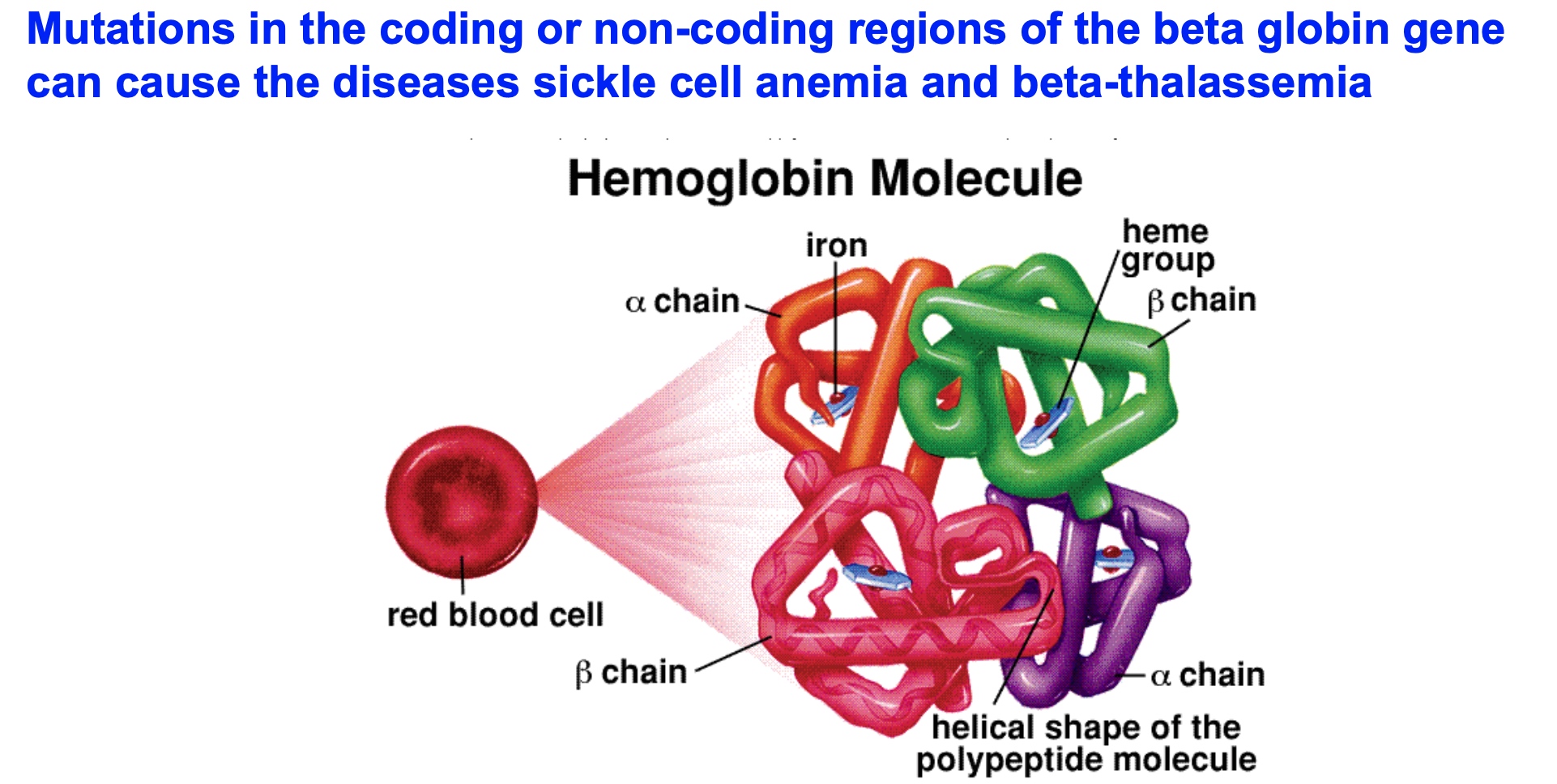
Case Study: Splicing and Human Disease
Part 1
Two patients come to you, presenting with severe anemia. You draw a blood sample and look at the cells under a microscope.

Anemia:
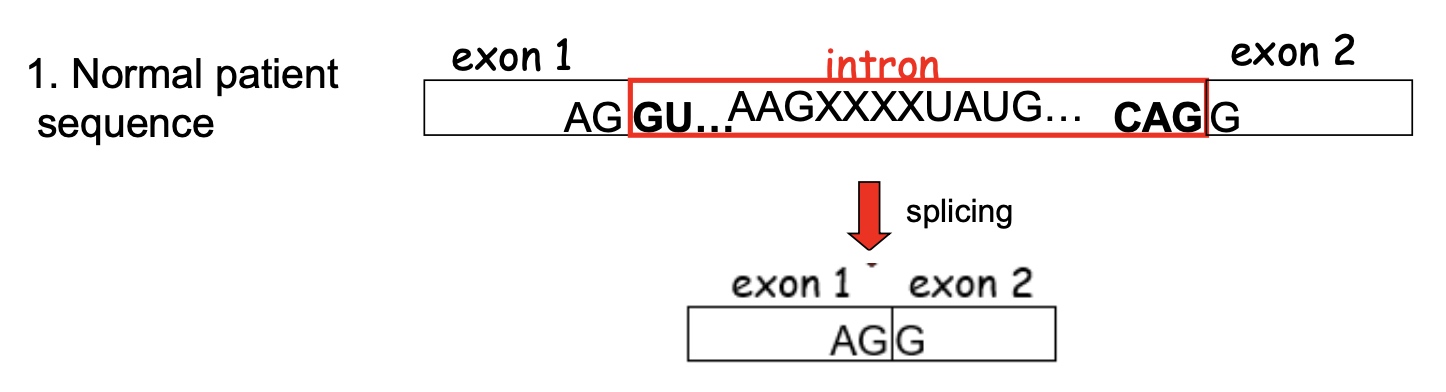
Part 2
You sequence the beta globin genes from a normal patient and the two affected individuals. Below is shown the sequence for the beta globin gene for a normal patient.

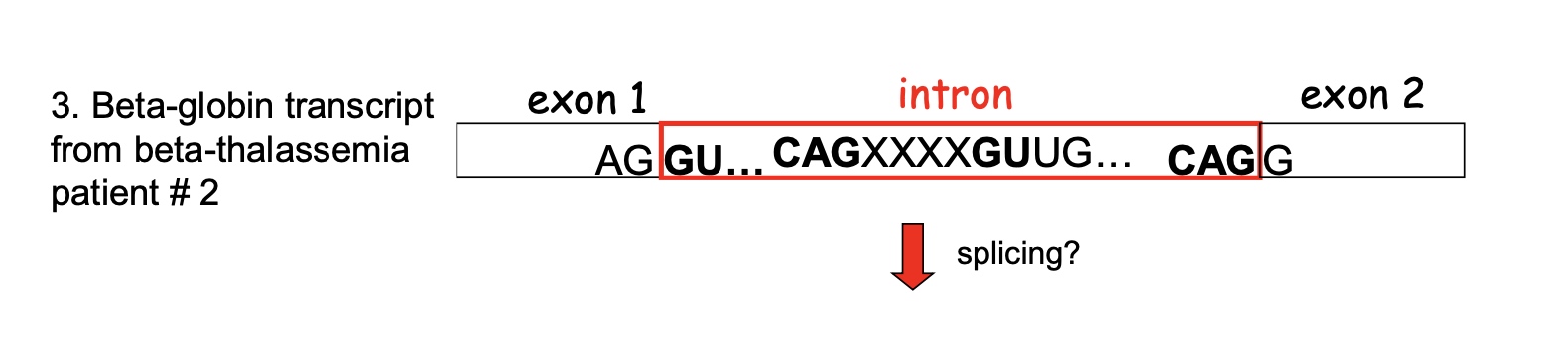
In the patient #2, the XXXX will also be in exons.
Part 3
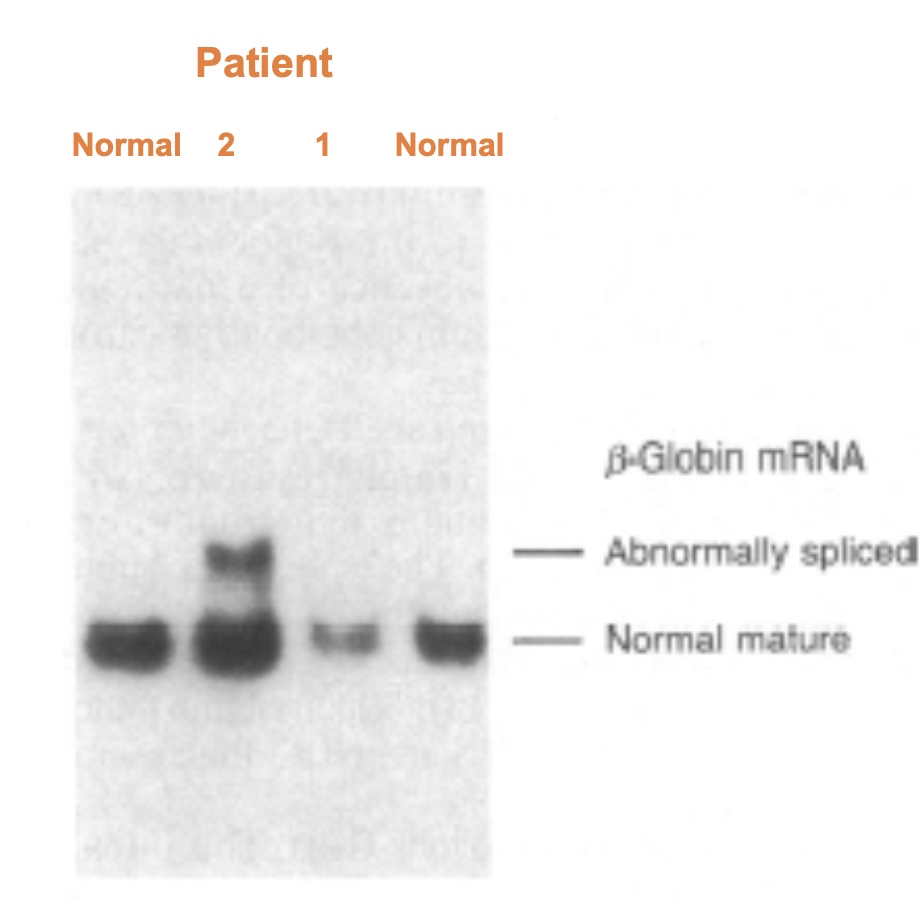
This is a northern blot showing the β-globin mRNAs from each patient.
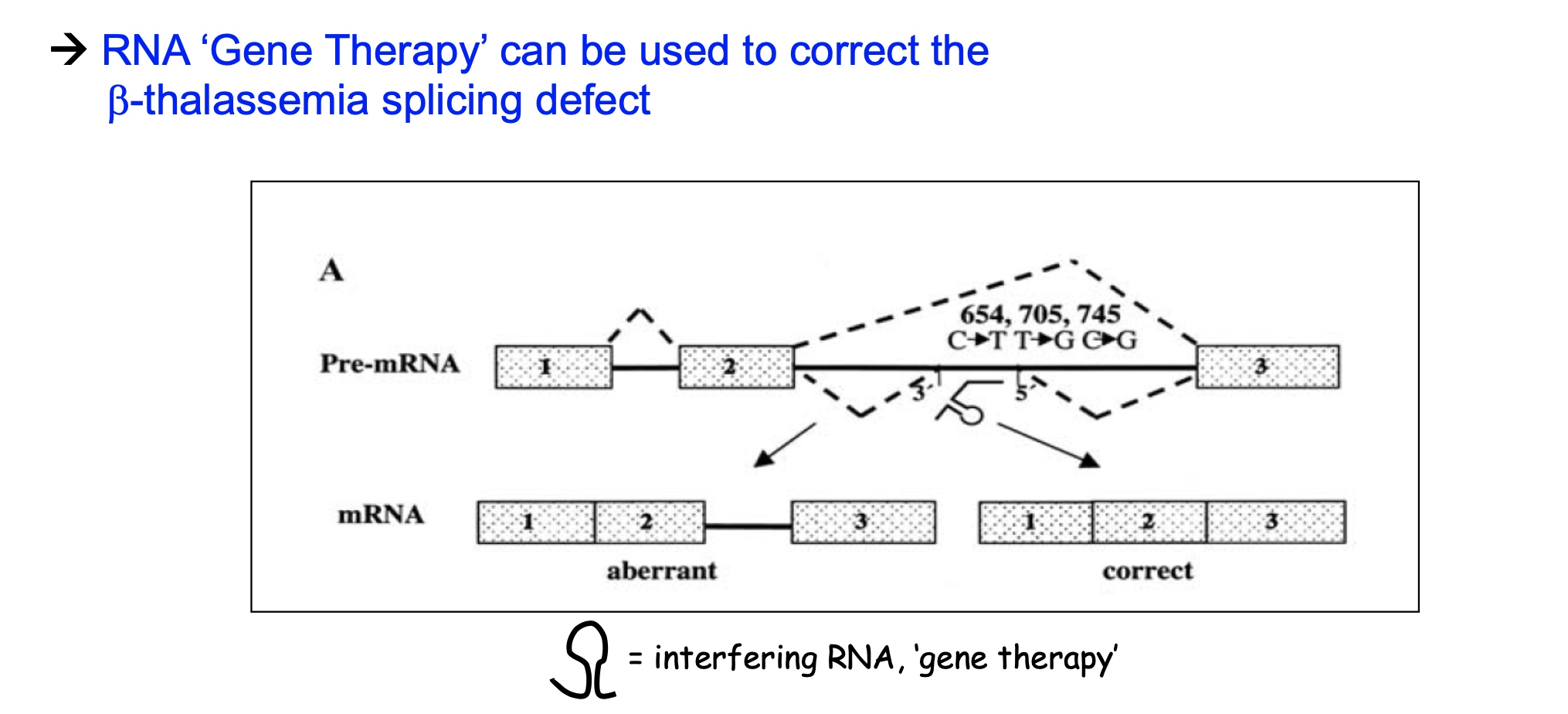
A novel therapeutic for treatment of beta-thalassemia: