How is Splicing Fidelity Regulated? SR Proteins Recruit Splicesome Components
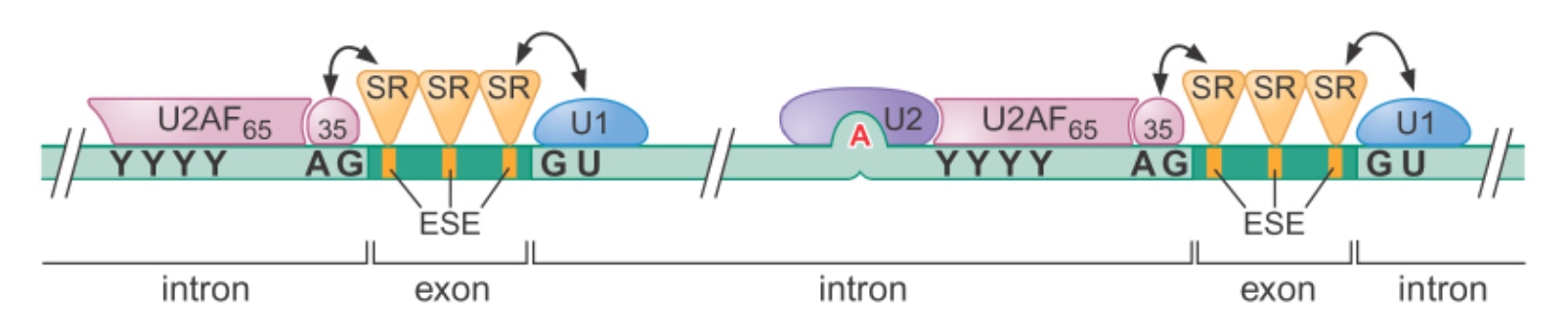
- SR (serine/arginine rich) proteins bind to ESE (exonic splicing enhancer) sequences
- They help to position the snRNPs correctly
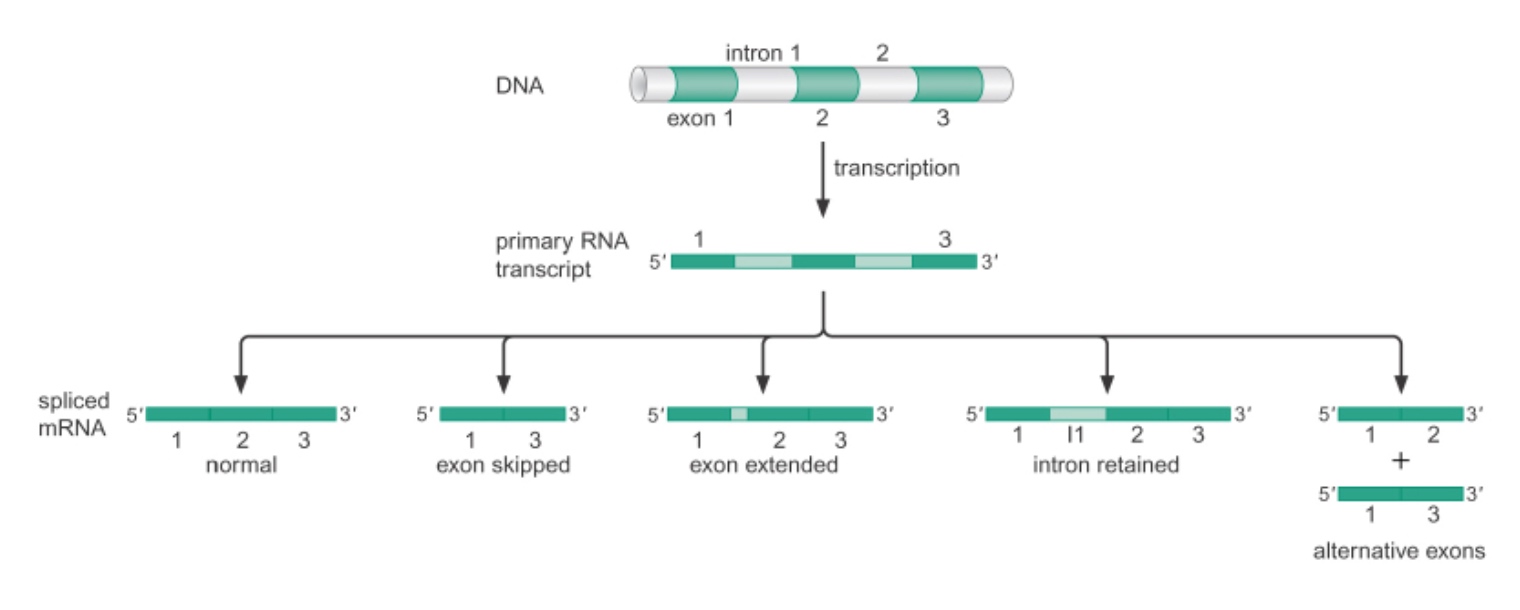
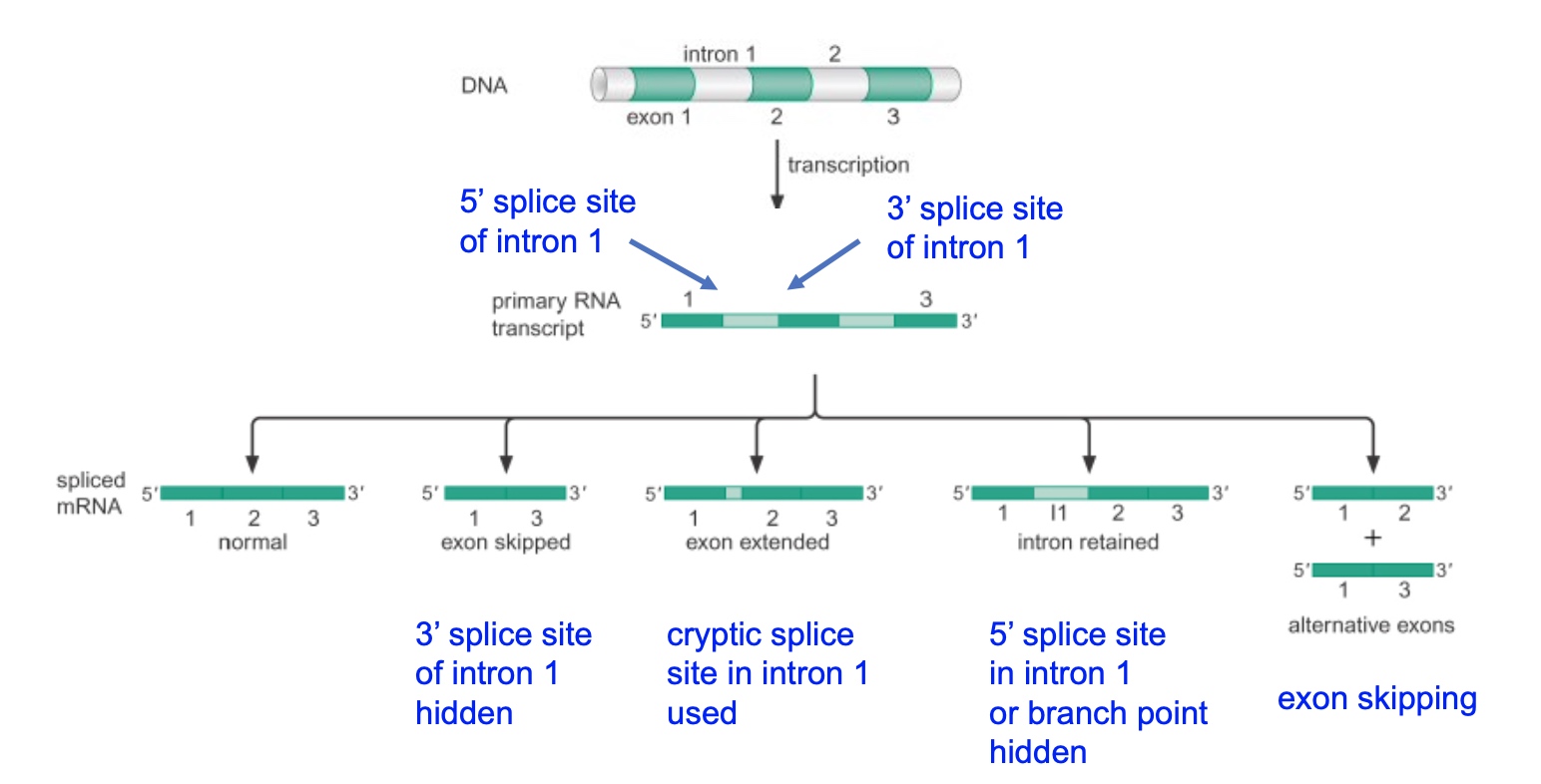
Alternative Splicing
Take in the same information and translated it into different products
ss: Splice sites(in the intron)
Poll Everywhere Question
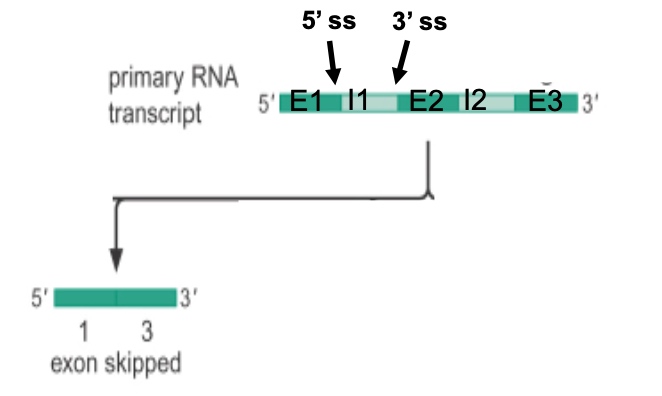
A mutation in which of these splice sites could result in the skipping of Exon 2?
- A. 5’ splice site of intron 1
- B. 5’ splice site of intron 2
- C. 3’ splice site of intron 1
- D. 3’ splice site of intron 2
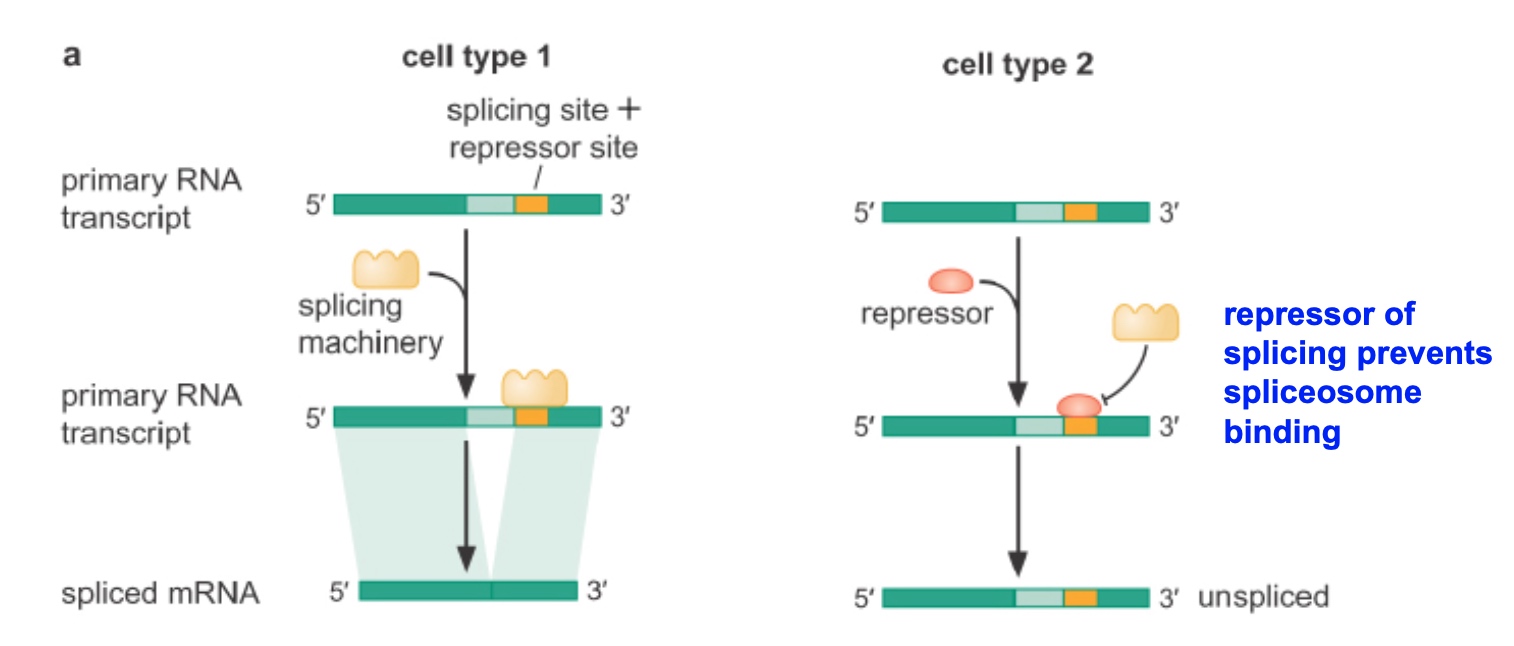
Alternative splicing can be regulated by splicing repressors
Splicing Repressor
- Splicing Repressor: repressor of splicing prevents spliceosome binding
Splicing Enhancer
- Activator of splicing promotes spliceosome binidng.
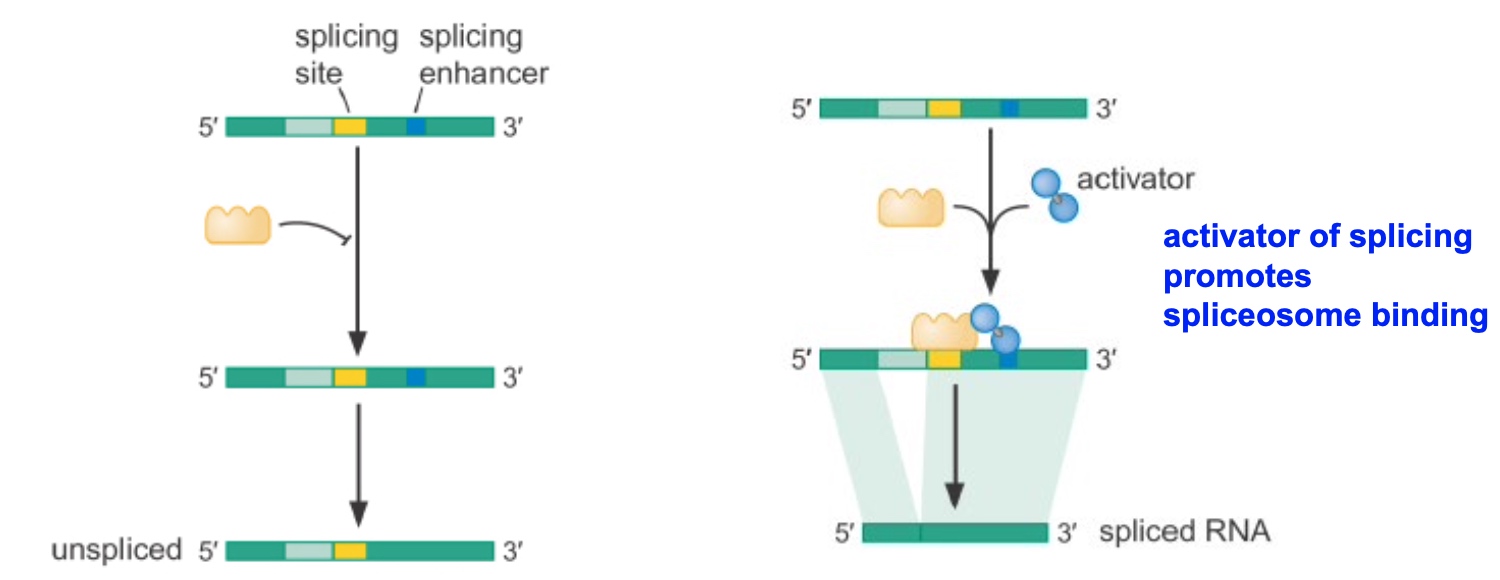
Both splicing repressors and enhancers are important during fruit fly sex determination!
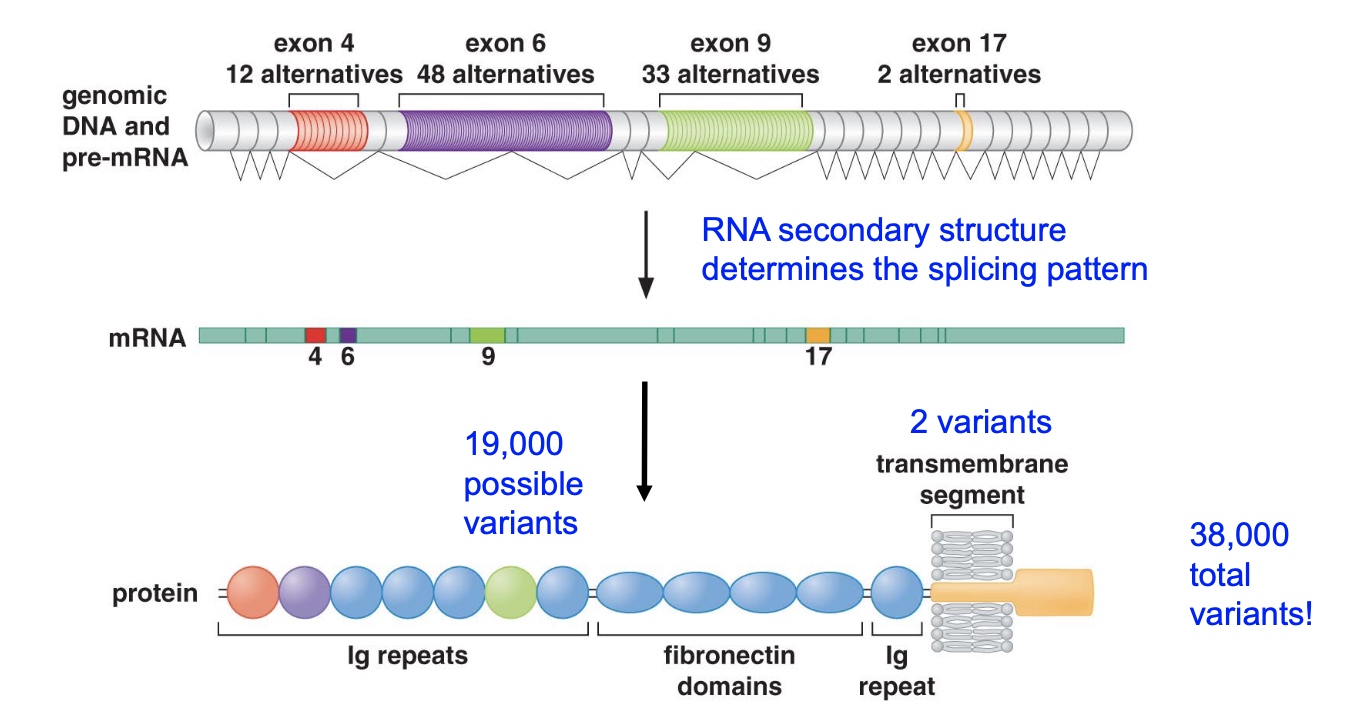
Example # 1: The Drosophila DSCAM gene Alternative splicing to the extreme
DSCAM is involved in innate immunity and neural patterning.
Example # 2: Alternative processing of the rat calcitonin gene transcript
CGRP: Calcitonin Gene-Related Peptide
Splicing skip on 4.
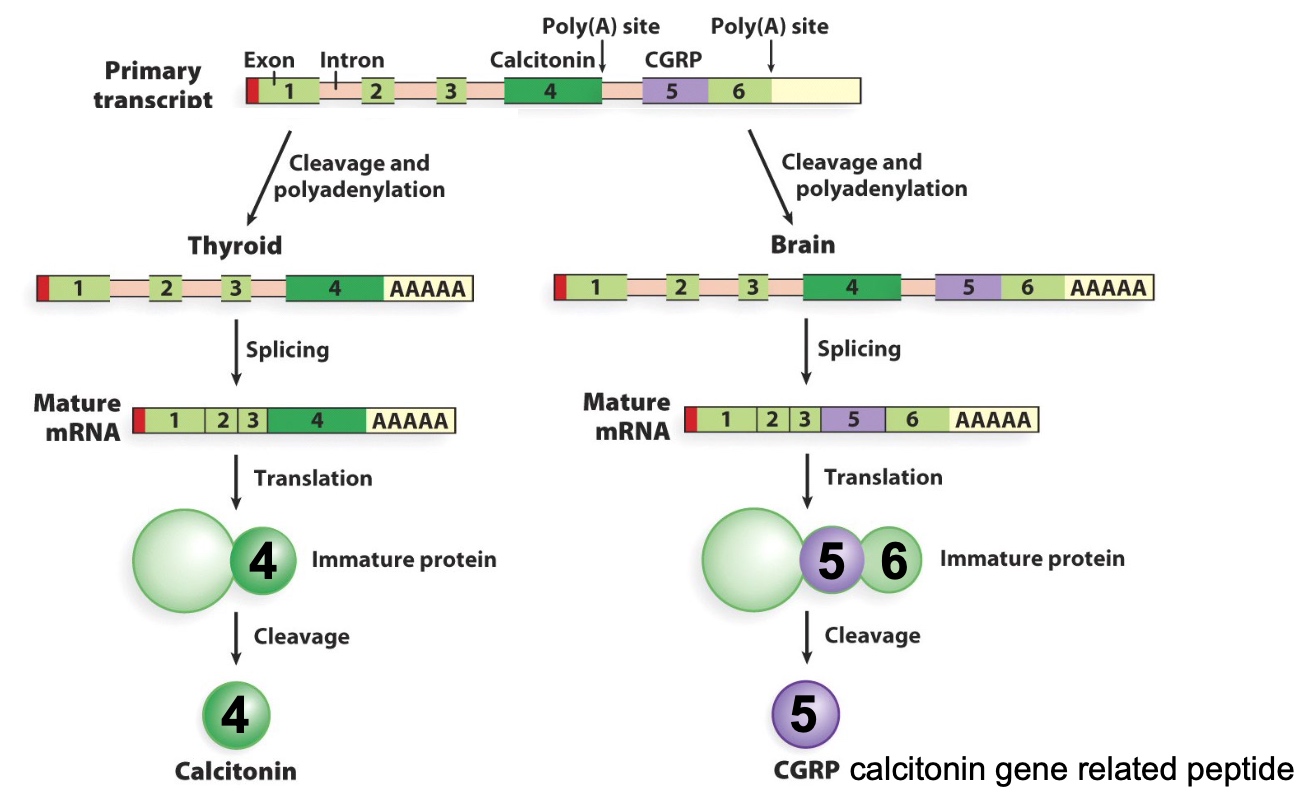
The poly A tail will not be added until the the mRNA strand is cut at the poly A tail.
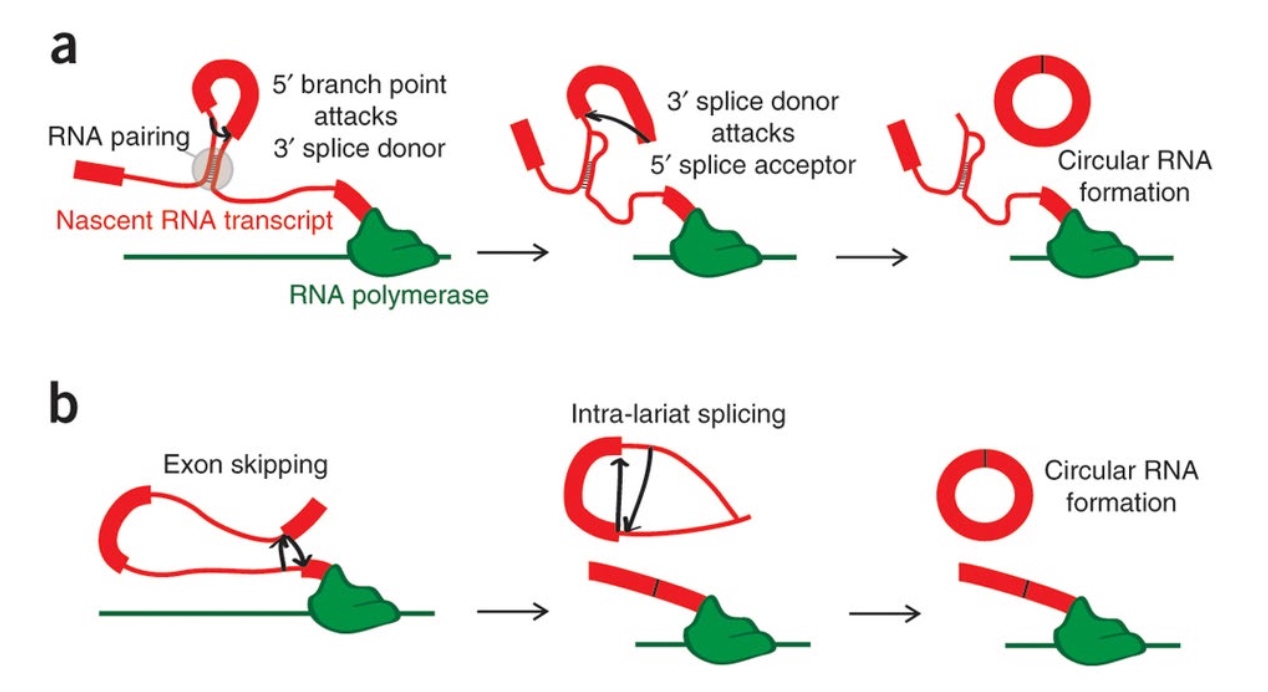
Backsplicing can Create Circular RNAs(circRNAs)
circRNAs are being implicated in many human cancers
circRNAs are common and have important functions in cells
Functions: regulate gene expression some are translated into proteins
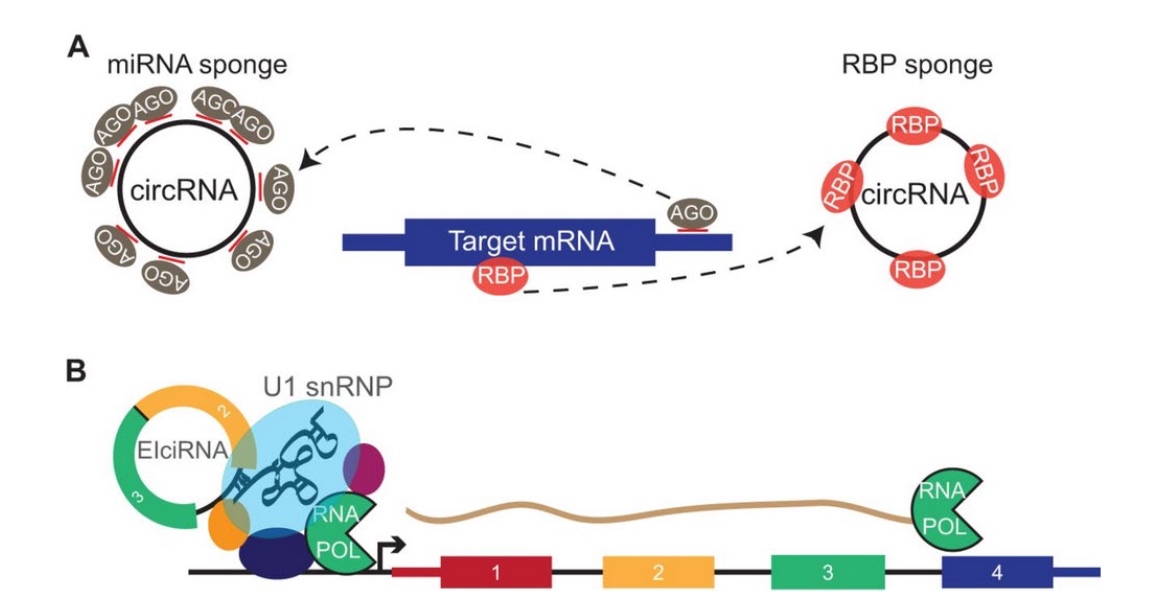
circRNA can be an RNA sponge / work with spliceing factor to do splicing.
Poll Everywhere
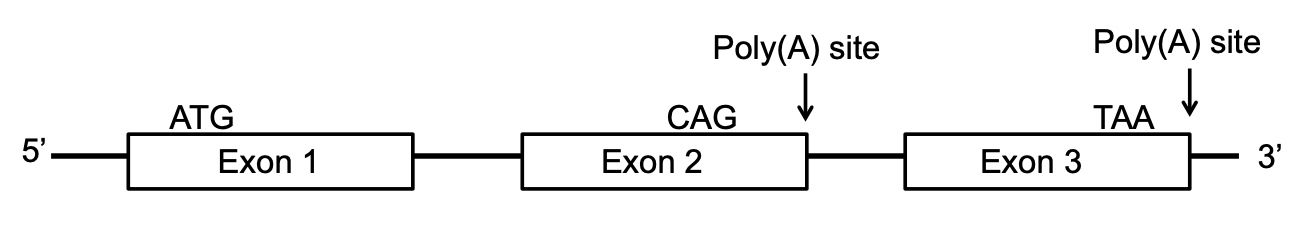
Which of the following could result in a PROTEIN containing exon 3 coding sequence?
- A. normal linear splicing
- B. alternative splicing (exon 2 skipping)
- C. cytosine deamination of the C in exon 2
- D. backsplicing to produce an exon 2 circRNA
- E. mutation of the first Poly(A) site
TAA is the stop site.
cytosine deamination of the C will cause C to U. UAG is a stop codon.
backsplicing will move Exon2 out, which will produce normal Exon3.
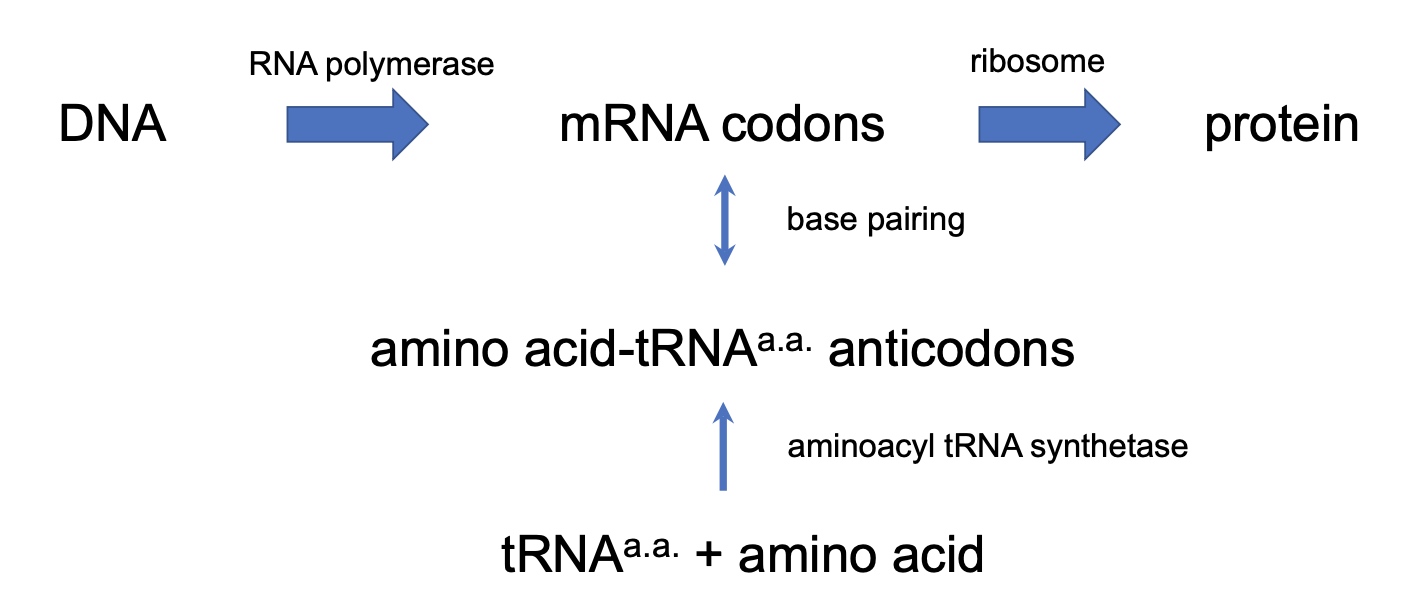
Interpreting the Genetic Code
Overview of Protein Translation
Poll Everywhere Application Question
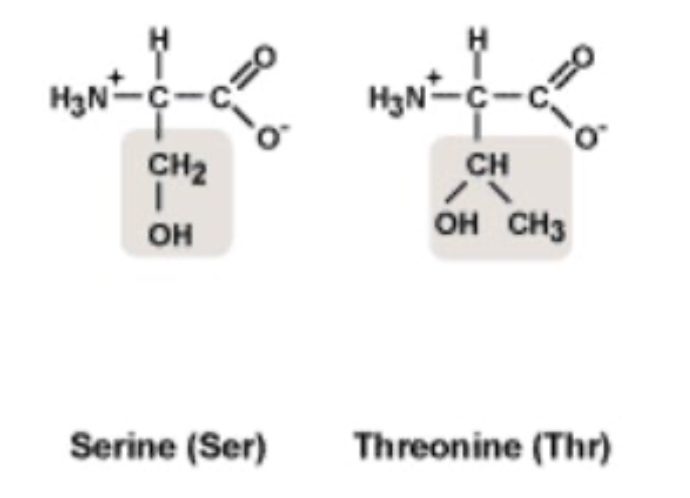
Given the amino acid structures below, which of these two tRNA synthetases is likely to have proofreading activity?
- A. seryl tRNA synthetase
- B. threonyl tRNA synthetase
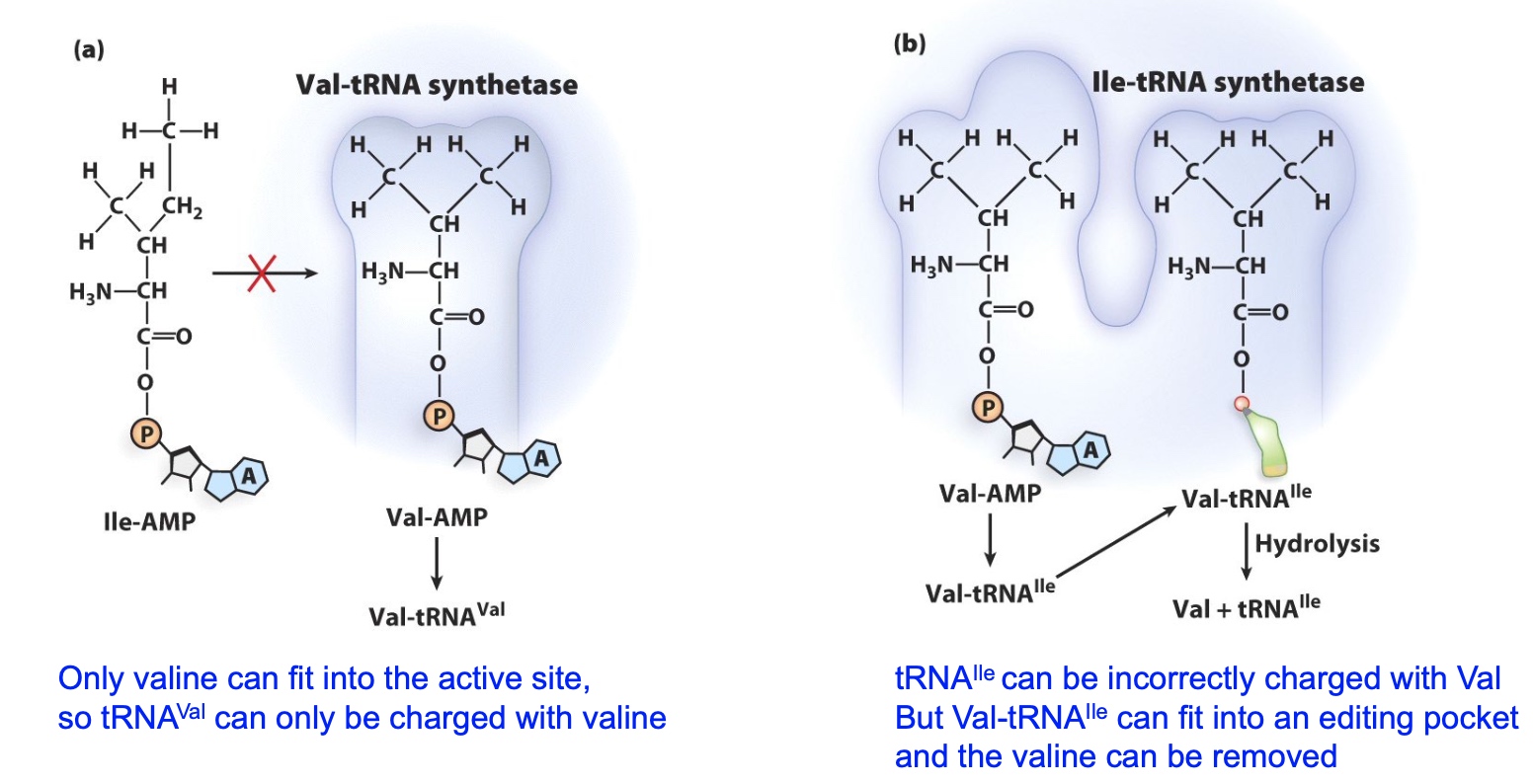
Aminoacyl-tRNA synthetases have proofreading capacity
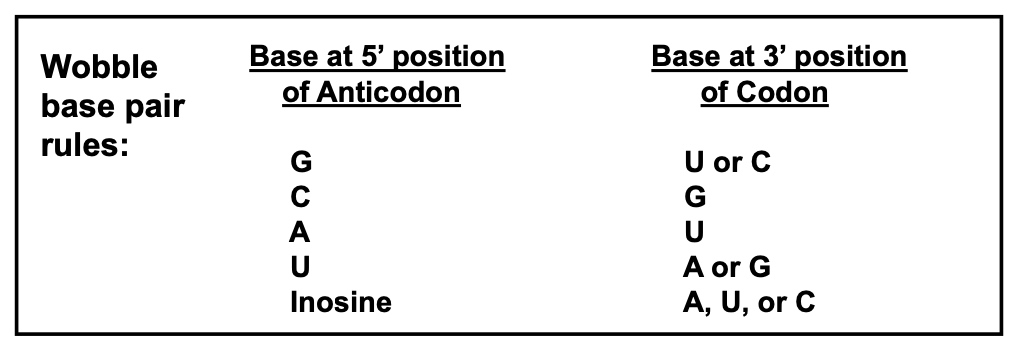
Wobble Rules (Wobble base pairing decreases the total number of tRNAs)
Because of the wobble effect, there are not 61 different tRNAs
There are ≥ 32 tRNAs, depending on the organism
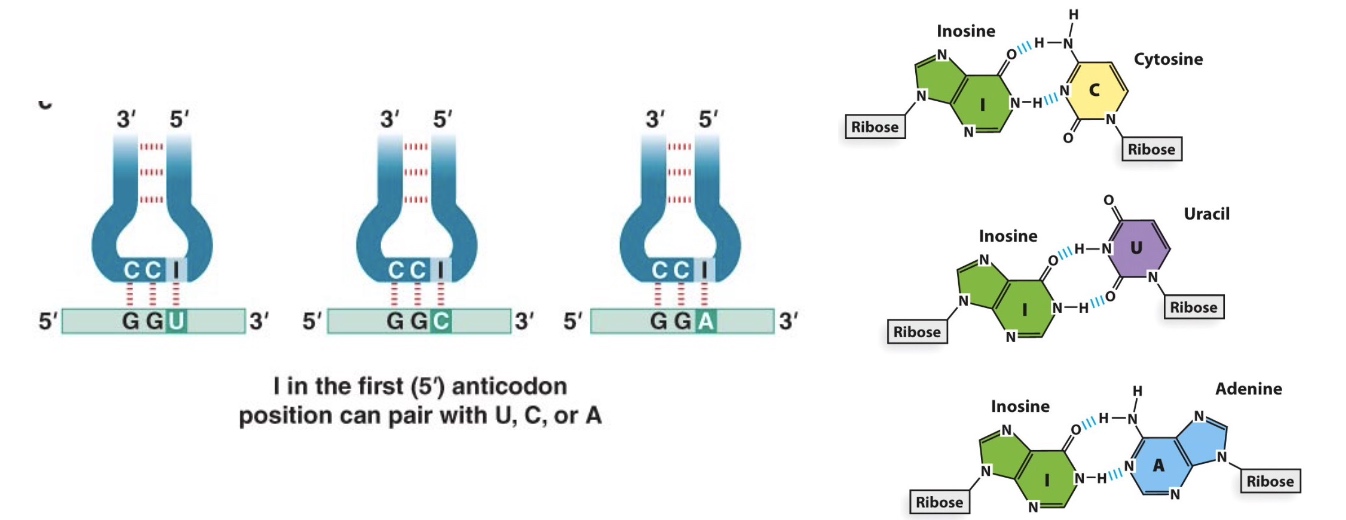
Poll Everywhere application question:
Given the wobble rules, what is the minimum number of tRNAs and tRNA synthetases that are needed to recognize all possible codons for the amino acid R?
- A. 2/1
- B. 3/1
- C. 2/2
- D. 3/2
- E. 3/3